Projects
Sometimes I code and I like it and sometimes I try to publish it too…
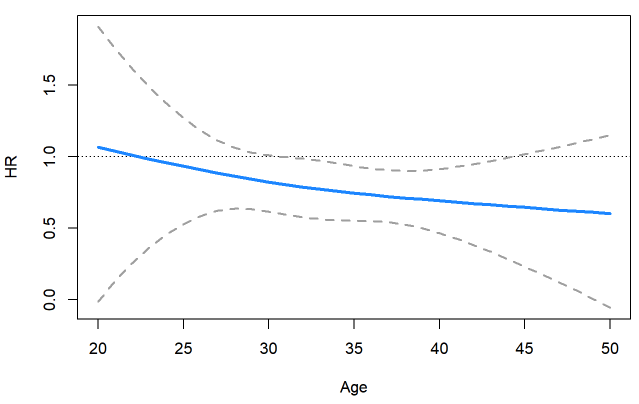
interactionHR
interactionHR is an R package to estimate hazard ratios
of a variable change when interacting with another variable.
Simple interaction Cox models and restricted cubic spline are implemented.
Confidence interval can be calculated via delta method or bootstrap.
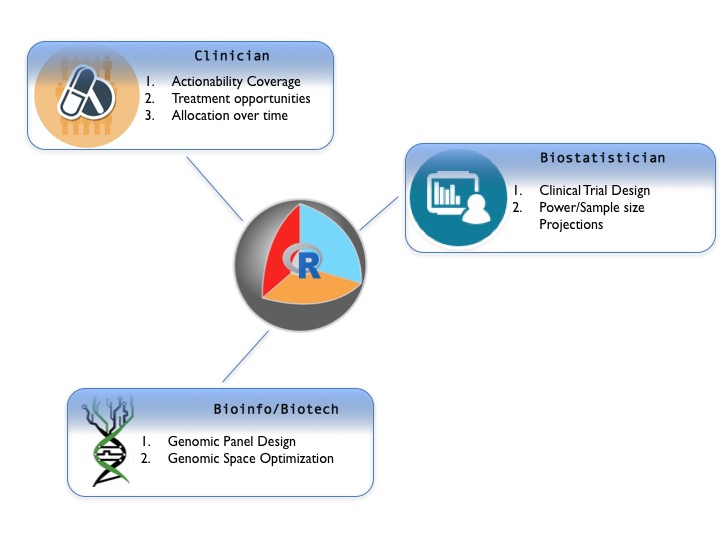
Precision Trial Drawer
Available on Bioconductor!
Full documentation and examples on the PTD Official Page
Melloni GE, Guida A, Curigliano G, Botteri E, Esposito A, Kamal M, Le Torneau C, Riva L, Magi A, de Maria R, Pelicci P, Mazzarella L (2018). Precision Trial Drawer, a Computational Tool to Assist Planning of Genomics-Driven Trials in Oncology JCO Precision Oncology. doi: 10.1200/PO.18.00015.
PTD provides a series of tools that can help bioinformaticians,
clinicians and biostatisticians to design, analyze and finalize
custom gene panels for cancer genomics clinical trials.
LowMACA
LowMACA is a method able to assess specific characteristics of rarely
mutated genes that show patterns of positive selection by aggregating
mutational patterns of several genes that have a
high level of sequence similarity
Available on Bioconductor!
Melloni GE, de Pretis S, Riva L, Pelizzola M, Céol A, Costanza J, Müller H, Zammataro L LowMACA: exploiting protein family analysis for the identification of rare driver mutations in cancer BMC Bioinformatics. 2016 Feb 9;17(1):80. doi: 10.1186/s12859-016-0935-7
TransformPhenotype
A shiny app to normalize continuous traits in Genome-wide
association studies
Gilly A, Kuchenbaecker K, Southam L, Suveges D, Moore R, Melloni G, Hatzikotoulas K, Farmaki AE, Ritchie G, Schwartzentruber J, Danecek P, Kilian B, Pollard M, Ge X, Elding H, Astle W, Jiang T, Butterworth A, Soranzo N, Tsafantakis E, Karaleftheri M, Dedoussis G, Zeggini E Very low depth whole genome sequencing in complex trait association studies, bioRxiv (2017) doi: https://doi.org/10.1101/169789 )
DOTS-Finder
Driver Oncogenes and Tumor Suppressors finder is a tool
to identify driver genes from exome-sequencing data
from a cohort of cancer samples.
Available on SourceForge
Melloni GEM, Ogier AGE, de Pretis S, Mazzarella L, Pelizzola M, Pelicci PG and Riva L. DOTS-Finder: a comprehensive tool for assessing driver genes in cancer genomes. Genome Medicine 2014, 6:44, DOI: 10.1186/gm563